Machine learning for materials and molecules: toward the exascale
PI: Michele Ceriotti (EPFL)
Co-PIs: Guillaume Fraux
July 1, 2021 – June 30, 2023
Project Summary
The past decade has seen the rise of machine learning techniques in all fields of science. The impact of these techniques has been particularly substantial in computational chemistry and materials science, and in general in the atomic-scale modeling of matter. Data-driven approaches helped address several of the outstanding challenges in the field: first, the automatic classification and dimensionality reduction made it possible to understand simulations with increasing size and complexity. Secondly, fitting general potentials — trained to reproduce accurate electronic structure calculations — opened the way for simulations that recreate realistic thermodynamic conditions, without giving up the precision and the predictive nature of first-principles methods.
A crucial step in these applications involves mapping atomic configurations to a set of features. This step dramatically affects the accuracy of the model, because it encodes symmetries, sum rules, asymptotic tails, and other physical concepts that improve the data efficiency of the training exercise. Evaluating these representations is also usually computationally expensive, and for the simpler ML models it is by far the most demanding step of a ML workflow. The PI and his group has contributed a number of foundational insights on the nature, behavior, and evaluation cost of representations for atomistic ML. The fundamental understanding is that all representations currently in use can be regarded as differing in implementation but very similar in substance – essentially being equivalent to n-point correlations of the atomic density. Building on these insights, the group of the PI, in collaboration with the Laboratory of Multiscale Mechanics Modeling of EPFL and in the context of the NCCR MARVEL, has developed librascal, a library dedicated to the efficient evaluation of Representation for Atomic SCAle Learning.
Librascal is built around a set of clear design principles, inspired by those applied to the development of i-PI, a universal force engine for advanced molecular dynamics developed in the PI’s lab: (1) Code modularity – the implementation is broken into simple units that reflect the mathematical structure of the representation problem; (2) Reusability and integration – the code is designed to be easily combined with existing packages to perform auxiliary tasks (from the handling of atomic configurations to the evaluation of ML models); (3) Bleeding-edge features –the implementation effort goes hand-in-hand with more formal, conceptual developments, so that the library can quickly incorporate algorithmic advances; (4) Code quality – best development practices, such as the use of unit tests, continuous integration and code coverage monitoring are enforced, and a high-quality documentation is maintained to simplify contributions and usage.
Librascal has already evolved to the level where it is used as the primary tool for ML applications in our laboratory, and it is making unique features available, such as covariant descriptors to predict tensorial properties. This far, however, it has been developed primarily as a serial library, focusing on features and on establishing a sustainable infrastructure for the code. The purpose of this PASC proposal is to make librascal ready for exascale applications, implementing node-level parallelism, porting the most demanding sections of the code to the GPU, improving the integration with efficient ML libraries and molecular dynamics engines, and maximising ease of use.
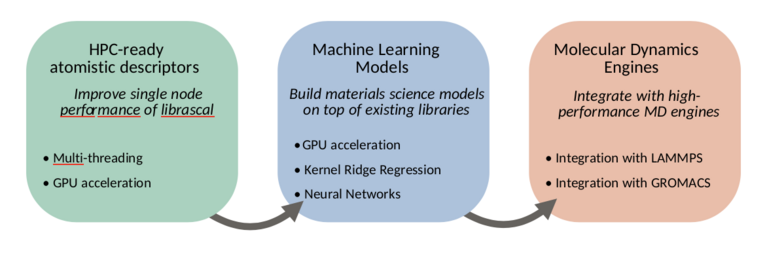
To this end, we will work in three main directions, summarized in figure 1: improving the node-level performance of librascal, including the development of GPU-accelerated feature evaluation, adding integration with machine learning libraries to allow accelerated model evaluation, and integrating librascal and the machine learning models within existing, high-performance molecular dynamics engines.
HPC-ready atomistic descriptors We will adapt the descriptors computation in librascal to HPC environments. Of the three main level of performance improvement typically used in an HPC setting (distributed parallelism, shared memory parallelism and GPU/coprocessor acceleration), distributed parallelism is already taken into account by the molecular dynamics engine using MPI and a spatial decomposition algorithm (see figure 4). As long as the descriptor is local, i.e. depending only on the position of atoms close to the one being considered, distributed parallelism is already available. For this reason, we will focus our effort on improving the single-node performance of librascal, by adding multi-threading and where possible GPU acceleration to the descriptor calculation.
Integration of models Currently, librascal implements some models, such as Gaussian process regression. A more sustainable approach is to ensure that librascal exposes an API that allows integration with standard ML libraries, such as PyTorch. We will build a library of wrappers adapted to the prediction of materials and molecular properties; in particular the energy, forces and virial acting on an atomic scale system, but also the tensorial properties that are a distinctive feature of librascal. We will structure the implementation to leverage the existing GPU-accelerated models in PyTorch or similar libraries.
Integration with MD engines Finally, we will integrate librascal and the ML models library with existing molecular dynamics engine. In particular, we envisage focusing on LAMMPS as an engine that is widely used for materials science applications, and GROMACS as one that is suitable for the simulation of biological systems.
The synergy with other methodological and materials discovery effort within the group of the PI and the NCCR MARVEL at large will provide impactful scientific applications, give opportunities to demonstrate the HPC capabilities of librascal on CSCS and european hardware and ensure its sustained development after the end of the present project.